Choosing cell modelling software: Virtual Cell, Cytoscape, CellDesigner, E-Cell
10th May 2007
I’m planning to reconstruct (based on literature and some original research) a specific cellular regulatory network. For this I decided to use some specialized biological modelling software. The requirements I had were pretty simple:
- must have SBML support. SBML appears de-facto standard for biological model notation;
- must be fairly frequently updated;
- should be feature-packed and easy to use. However, this requirement can only be checked after some use, and I was pre-selecting, not reviewing.
Software put into the title of the post was found to be the most mature and interesting from the usage perspective. However, there are more than those mentioned software tools reviewed. Reviews are based primarily on the information from official websites and documentation; some tools (like VirtualCell) are reviewed somewhat more thoroughly.
I found a presentation, which lists some of the cell modelling tools to start with (and also has a comparison table, but it’s too outdated).
Update: after 40% of the post was written, I came across the Computational Cell Biology book’s website, which has a links section, with a list of cell modelling software.
Update 2: one more list.
VirtualCell
First I tried VirtualCell, version 4.2. It is an online tool, which I consider to be a drawback, as I may choose to read some article (and extend the model) somewhere far from civilization, where the most I can get is a 220V plug, and no scent of internet at all.
VCell supports a number of different model types, as seen from the ‘new model’ dialogue (composite image):
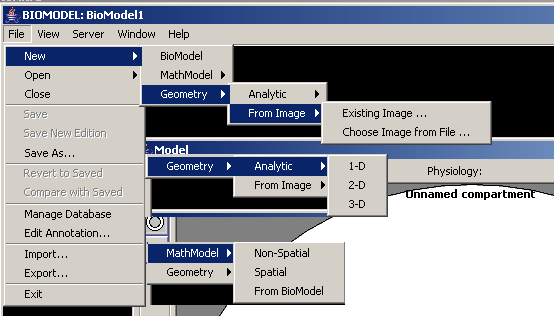
As you can see, three main types are BioModel, MathModel, and Geometry, which appears to cover most of the needs bio-modeller may have.
Any models are stored and simulated on the server; Java client appears to be used for visualization and parameters/geometries editing only.
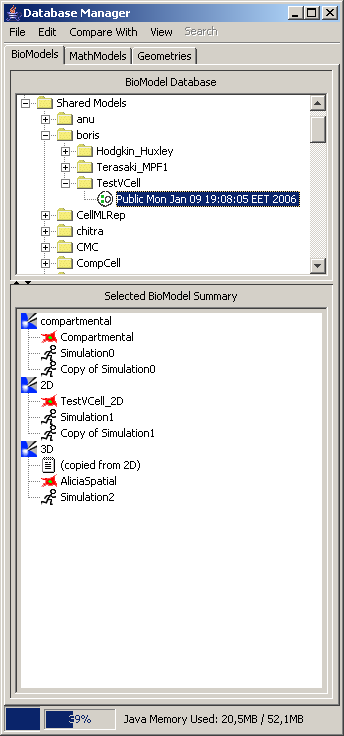
Each model may have several different-type components (i.e., a model may start as bio-model, then be extended with geometry, and then – with math model). When we open a sample model, we get the window like this:
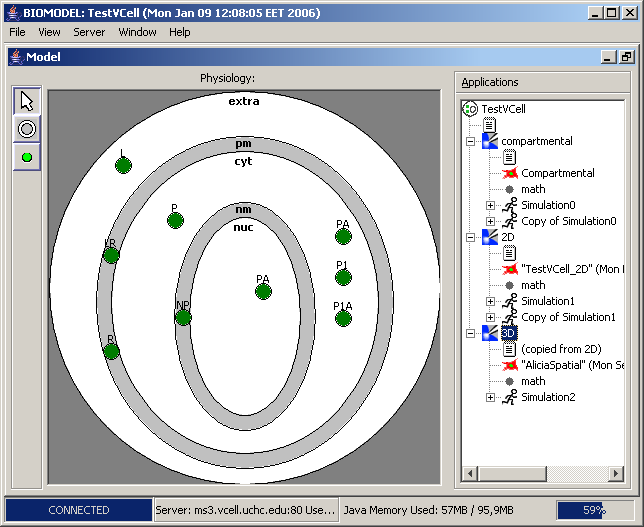
Next screenshot shows this model’s parameters:

Virtual cell 4.2 supports import from XML files, and a number of formats for export:
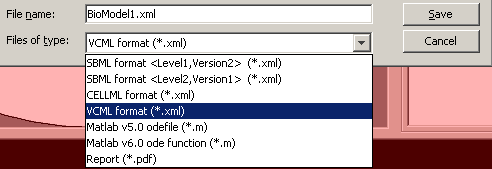
In conclusion, Virtual Cell made a good impression. It is a well thought-through software, versatile enough to satisfy the demands I can currently imagine :). The single but large drawback, in my opinion, is the requirement for internet connection to work with it. Addition of some kind of a “permanent cache” (probably like the one found in MS Office 2007 Groove), which would allow to work on the model offline, and then synchronize with the server, might be beneficial.
(There is also a minor “drawback” in that VCell is programmed in Java, and I organically dislike memory/CPU-inefficient interpreted-language solutions for being slow and excessively memory-consuming. But nowadays Java is so popular, that I’m just forced to silently accept software, written in Java – if there is no alternative, of course.)
NetBuilder
NetBuilder is a “graphical tool for building logical representations of genetic regulatory networks”. NetBuilder is based on the concept of constructing gene-focused regulatory networks like circuit boards; in the “Genome” view (also “design mode”) you add separate blocks – genes – and specify how they (with the intermediary products help) influence each other. Then in the “Nucleus” (simulation) view you can see how your model behaves. The program is simple, but so far seems the most convenient for the genetic regulatory networks evaluation. There is an interesting comparison of the model-predicted and actually-measured expression values for the Endo16 gene: first see the model, then see simulation results.
Unfortunately, the latest version of NetBuilder (0.94) was released back in mid-2003. What a pity, really. Given SBML support, I might have chosen even the 0.94 version. But there is no SBML support there.
NetBuilder’
However, there is NetBuilder’ – this time in Python, not C++ and MFC. The project appears to be inspired by Maria Schilstra. There is a short intro into netBuilder’, and a sourceforge project page (at the moment of writing, there is nothing to download from there – “No File Packages Defined”).
NetBuilder’ is a modelling and simulation environment that is specifically aimed at the study of genetic regulatory networks. Therefore its mathematical modelling capabilities (definition of the kinetic laws) are limited in comparison with those in tools like Gepasi/COPASI, Jarnac, etc. The differences will be discussed in more detail below. Its simulation capabilities are the
same.
As I need software now, and I didn’t come across any kind of public development roadmap for NetBuilder’, I must drop it as an option from my review.
Jarnac, JDesigner, Systems Biology Workbench form the whole package for biological modelling and simulation. It was last updated in November, 2006 (at the moment of writing), and is supported by DARPA, NSF, DOE Genomics etc. Systems Biology Workbench is intended as an open-source framework to connect heterogeneous software from the cell modelling field. JDesigner 1.980 looks to me as a metabolic pathway modeller, which is not exactly what I want (I need genes as well). Actually, among the sample models there are models with genes and their products, but so far I feel like something with a better interface… Will see what’s else except for SBW, but it looks solid and worth trying. (Note: it appears that EBI is cooperating with SBW developers, which isn’t bad as well.)
Cellerator
Cellerator was designed to simulate
1. signal transduction networks (STNs);
2. cells that are represented by interacting signal transduction networks; and
3. multi-cellular tissues that are represented by interacting networks of cells that may themselves contain internal STNs.
By this description Cellerator fits my requirements even better than I could imagine starting the search! Except for: it’s Mathematica extension (thus one needs Mathematica, which isn’t free), and it’s heavily modelling-oriented: converts any relations into ODE (thus inexact data isn’t welcome). There’s GUI for Cellerator, called Sigmoid Model Explorer. Cellerator is not developed any more – only bug fixing is done – so it’s not worth any further review. The new product is called xCellerator.
xCellerator
xCellerator is also a Mathematica package. However, it’s index page has “This page was last updated 13 August 2005″ note in it, which is too bad for such a promising product. Note, that available version is only “alpha”, which means “early development”. The project might be reborn someday (or may totally die silently), but anyway it won’t be able to compete with now-established products due to the large time-gap.
Next one in the queue is Cellware. However, it’s latest version 3.0.2 was released back on 31-03-2005. Dump and move further.
(It appears to me that majority of afterwards-dying software tools for science are produced by scientists. Tools die just because the field of interest changes, and there’s nobody to support the software. Unlike science-produced tools, commercial have the reason to survive, and seem to be doing overall better than pure-science-derived products. Correct me if I’m wrong.)
Gepasi is even older that the previous one.
(Am I wrong, assuming that successful software is regularly updated? What if _ALL_ features are implemented, and further changes can only change the UI? Windows XP vs Windows Vista comes to memory here. But I’m right: SBML is being developed and changed, and programs supposed to support it must at least be updated to account for SBML changes. Thus older releases mean worse features support.)
There’s also DynaFit, which is a kinetic simulation tool. It was last updated in mid-2006, which is not bad. But it doesn’t fit my requirements and necessary scope.
Basically, only three options had drawn more of my attention: CellDesigner, Cytoscape, and E-Cell. Let’s have a more detailed look at those.
CellDesigner
CellDesignerTM is a modelling tool of biochemical networks with graphical user interface. It is designed to be SBW (Systems Biology Workbench) compliant, and supports SBML (Systems Biology Markup Language) format.
Last updated: January 2007
CellDesigner appears closely related to the SBML consortium:
The development of this website is funded by the Systems Biology Institute, and Kitano Symbiotic Systems Project, ERATO-SORST, JST. We wish to help you enter this exciting field.
CellDesignerTM is a structured diagram editor for drawing gene-regulatory and biochemical networks.
Networks are drawn based on the process diagram, with the graphical notation system proposed by Kitano, and are stored using the Systems Biology Markup Language (SBML), a standard for representing models of biochemical and gene-regulatory networks. They are able to link with simulation and other analysis packages through Systems Biology Workbench (SBW).
CellDesigner has deeply thought-through Kitano graphical notation. Probably even too detailed for my initial purposes. It is convenient for modelling, might as well work fine for simulations and analysis.
There is another tool, Complex Pathway Simulator (COPASI), which is a software application for simulation and analysis of biochemical networks. The “news” is that COPASI and CellDesigner announced their collaboration that aims at integrating the two tools. This integration will allow users to edit their system of interest using the advanced graphical interface of CellDesigner and analyse it’s properties with the powerful methods provided by COPASI in a seamless fashion. The collaboration is supported by a grant by the German Ministry of Education and Research.
COPASI, based on my impressions, is for simulation and analysis (it is like like E-Cell). COPASI is definitely good, and more user-friendly than E-Cell (based on comparing the manuals of the two). However, COPASI itself was not suitable for modelling (no model builder). With the forthcoming integration with CellDesigner, those two tools may become a kind of an “ultimate solution”.
Cytoscape
Cytoscape is a Java tool.
Last updated: January 2007.
Project started in 2002.
Primary uses of Cytoscape: data management, data organization, data collection and data visualization.
Analysis with Cytoscape is limited to plugins. Simulation is even more limited.
My verdict: good for data pulling from databases, visualization and data management (well, I’m repeating official information). Cytoscape might be good to start with, thanks to simplicity and nice GUI.
E-Cell Project
E-Cell project is an international research project aiming to model and reconstruct biological phenomena in silico, and developing necessary theoretical supports, technologies and software platforms to allow precise whole cell simulation.
Last updated: mid-2006.
Project started: 1996 (the oldest among the reviewed tools?).
Written in C++/Python.
Supports SBML levels 1/2.
Supports distributed computing. (Distributed computing is running multiple simulation sessions on different computation nodes on cluster or grid environments.) Currently Sun Grid Engine and Globus toolkit are supported.
E-Cell supports multiple cells/organelles, albeit in a complicated but logical EM/EML file format. E-Cell appears to have no visual model builder.
Some screenshots:
http://unix.freshmeat.net/screenshots/2138/
http://sourceforge.net/project/screenshots.php?group_id=72485
E-Cell project held a workshop in February 2007, which means that the project is definitely alive ![]()
There is also a list of publications (1999-2007) related to E-Cell – some of them (judging by the title) are definitely worth reading.
My verdict: good for analysis and simulations, lacks only model builder.
Coming to the final conclusions…
There is no obligation to use a single software tool for all your needs. Often, several tools are needed – with quite clear functional differences. Thanks to SBML, models can be first built in e.g. Cytoscape, then run in E-Cell, visualized in Cytoscape again. VirtualCell appears to be an uncommon tool, as it does (try to) integrate both modelling and analysis. Also, there’s SBW, which is an “integrator tool” itself.
Overall, it looks like:
- cytoscape is good in gathering and unifying data from different sources,
- CellDesigner is good at making models,
- E-Cell is good for simulations and analysis.
All the three are regularly updated, have long history, and are standalone (to the extent possible).
VirtualCell is an integral solution, but it’s an online tool which cannot be used offline, thus avoided.
Final conclusions:
- Use Cytoscape to try pulling data from different sources together, and for data visualization.
- Use CellDesigner to create models. CellDesigner can also be used for simulations and analysis with external (SBW-connected) tools, but this requires further investigation. Also, in the nearest future one may expect integration of CellDesigner and COPASI, which will bring together two high-quality software tools – one for building models, and another for analysis and simulations.
- E-Cell can be used for simulations and analysis. However, additional efforts must be put into learning E-Cell’s features to be sure if using E-Cell provides any advantages over using SBW-connected modules via CellDesigner.
I hope you found this information useful. If you notice any errors, or have important new information – I will be glad to see you commenting on this post ![]()

January 24th, 2009 at 3:09
[...] tutorial was also good, and referring to my older post on choosing cell modelling software, it is clear that CellDesigner is getting better with the new versions; it does include SBW [...]
December 16th, 2009 at 20:30
hi everyone,
I have an observation about Cytoscape. I was loading a network handly and when he got to 500 nodes, file is broken.
I have tried by all means, save the file. I had no luck. I wrote to the developers of Cytoscape and I have not had a response satisfactory outcome.
I think it is an uncommon event. but nobody gives you aid or solution when Cytoscape files are corrupted.
regards,
arsenio