25th February 2011
This overview presentation is two years old, but still a highly valuable resource: modules and tools mentioned are alive and useful.
I think this is the second presentation by Giovanni I’m embedding (first one being about GNU/make for bioinformatics).
Posted in Bioinformatics, Links, Python, Software | No Comments »
16th February 2011
Imagine you need to install pycassa (which uses easy_install). Here are the 2 (at maximum) very simple steps to have it properly debianized and installed on your Debian/Ubuntu:
- if you don’t have the python-stdeb package: sudo aptitude install python-stdeb
- pypi-install pycassa
That’s it.
Refer to stdeb readme for more information. You will need that if there are dependencies – which might not be resolved automatically by stdeb.
Before stdeb, it wasn’t exactly trivial to make a .deb from python module.
Posted in *nix, how-to, Notepad, Python, Software | 1 Comment »
25th January 2011
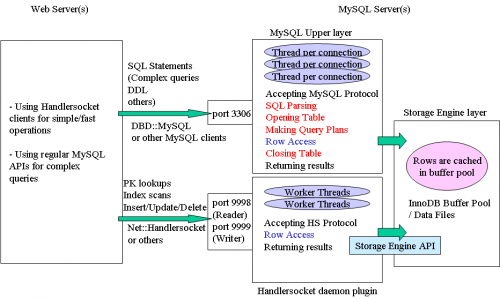
HandlerSocket provides a direct access to InnoDB storage, bypassing SQL interpretation layer. With in-RAM data, it may raise MySQL performance to 750000 queries per second.
Posted in Links, Software | No Comments »
10th January 2011
Updated on the 5th of March, 2010 (added flowdock and pivotal tracker, and also personal experience using a few of the previously described tools).
Back in 2007 I wrote a brief review of web-based project management tools. After that, I started using dotProject for personal projects management. I’m still using it, but for collaborative project management, communication, and tasks/milestones tracking dotProject isn’t perfect.
I need a tool, which is
- collaborative
- web-based (to allow effective collaboration)
- preferably free
- has concise per-project activity log
- minimal required functionality: tasks, milestones, files, and status updates.
After trying a few things, our small team settled for now on using github + pivotaltracker jira + confluence + flowdock.
Here’s a full list of tools briefly reviewed. I’ve been already using ProjectPier, so I’ll start with this software.
Read the rest of this entry »
Posted in Links, Software, Web | 11 Comments »
26th December 2010
On the 24th of September 2010 Microsoft posted a video showcase titled “A few perspectives on OpenOffice.org”. Here’s the page with the video: http://www.microsoft.com/showcase/en/US/details/faaf9eb8-77c6-4bed-bc08-c069a7bfbb04. It asks to install silverlight, and if you don’t want that – look for the Watch as WMV direct videostream link.
Just a single quote from Glyn Moody, Computerworld UK:
The criticisms made in the video are not really the point – they are mostly about OpenOffice.org not being a 100% clone of Microsoft Office, and compatibility problems with Microsoft’s proprietary formats. The key issue is the exactly the same as it was for the Mindcraft benchmarks. You don’t compare a rival’s product with your own if it is not comparable. And you don’t make this kind of attack video unless you are really, really worried about the growing success of a competitor.
See also what Savio Rodriguez (Infoworld) has to say about that video.
Posted in Misc, Software | No Comments »
4th December 2010
Here’s a simple and clear guide for gmail, which also definitely works with other relay hosts. I’ve used it to configure my ISP’s mail relay (they block outgoing port 25) on a Debian Squeeze laptop.
Posted in *nix, how-to, Links, Notepad, Software | No Comments »
16th November 2010
Imagine you need to get a few lines from a group of files with missing identifier mappings. I have a bunch of files with content similar to this one:
ENSRNOG00000018677 1368832_at 25233
ENSRNOG00000002079 1369102_at 25272
ENSRNOG00000043451 25353
ENSRNOG00000001527 1388013_at 25408
ENSRNOG00000007390 1389538_at 25493
In the example above I need ’25353′, which does not have corresponding affy_probeset_id in the 2nd column.
It is clear how to do that:
sort -u *_affy_ensembl.txt | grep -v '_at' | awk '{print $2}'
This outputs a column of required IDs (EntrezGene in this example):
116720
679845
309295
364867
298220
298221
25353
However, I need these IDs as a comma-separated list, not as newline-separated list.
There are several ways to achieve the desired result (only the last pipe commands differ):
sort -u *_affy_ensembl.txt | grep -v '_at' | awk '{print $2}' | gawk '$1=$1' ORS=', '
sort -u *_affy_ensembl.txt | grep -v '_at' | awk '{print $2}' | tr '\n' ','
sort -u *_affy_ensembl.txt | grep -v '_at' | awk '{print $2}' | sed ':a;N;$!ba;s/\n/, /g'
sort -u *_affy_ensembl.txt | grep -v '_at' | awk '{print $2}' | sed ':q;N;s/\n/, /g;t q'
sort -u *_affy_ensembl.txt | grep -v '_at' | awk '{print $2}' | paste -s -d ","
These solutions differ in efficiency and (slightly) in output. sed will read all the input into its buffer to replace newlines with other separators, so it might not be best for large files. tr might be the most efficient, but I haven’t tested that. paste will re-use delimiters, so you cannot really get comma-space “, ” separation with it.
Sources: linuxquestions 1 (explains used sed commands), linuxquestions 2, nixcraft.
Posted in *nix, Bioinformatics, how-to, Notepad, Software | 2 Comments »

