27th October 2010
- Install the annotationTools R package:
source(“http://bioconductor.org/biocLite.R”)
biocLite(“annotationTools”) - Download full HomoloGene data file from ftp://ftp.ncbi.nlm.nih.gov/pub/HomoloGene/current
- library(annotationTools)
- homologene = read.delim(“homologene.data”, header=FALSE)
- mygenes = read.table(“file with one entrez ID of the source organism per line.txt”)
- getHOMOLOG(unlist(mygenes), taxonomy_ID_of_target_organism, homologene) [alternatively, wrap the call to getHOMOLOG into unlist to get a vector]
It might be easier to achieve the same results with a Perl script calling NCBI’s e-utils.
Posted in Bioinformatics, how-to, Notepad | 2 Comments »
16th July 2010
In short: sudo mkswap -L new_swap_label -U old_swap_UUID /dev/sd_swap_device.
If you don’t care about the UUID: just sudo mkswap -L new_swap_label /dev/sd_swap_device.
Step-by-step:
Read the rest of this entry »
Posted in *nix, how-to | No Comments »
1st March 2010
There is an excellent step-by-step instruction on resetting the bad clusters counter of an NTFS partition with linux-ntfs tools. I’ve checked – it works as expected:
- Back up important data from partition just in case
- Find out size of ‘$Bad’ attribute in $Badclus using ntfsinfo -i 8 partition (partition is for example /dev/sda1). It will be the “Allocated size†value in the “Dumping attribute $DATA (0
x80)†(there will be two 0x80 attributes. Only one has an “Allocated size†line). Let us write down (remember) this size as ntfs_size. - Use ntfstruncate partition 8 0
x80 ‘$Bad’ 0 to set $Bad’s attribute length to zero. - Use ntfstruncate partition 8 0
x80 ‘$Bad’ ntfs_size to set $Bad’s attribute length back to proper value ntfs_size which was recorded in step 2. - Boot into Windows and run chkdsk -f diskname. It will find errors and should fix them.
However, Debian’s ntfsprogs package does not have the ntfstruncate binary.
Here’s how you can easily build one yourself (you may need a few extra packages with build tools for that):
Read the rest of this entry »
Posted in *nix, how-to, Software | 3 Comments »
27th October 2009
This query performs a table-wide search-and-repalce:
UPDATE `table_name` SET `table_field` = REPLACE(`table_field`,’string to search for and replace’,'replacement string’);
If you need a database-wide search-and-replace, you could try this script (I haven’t tested/used it myself).
Beware of the following gotchas:
- wrong query syntax may ruin the field you are performing replace on, so always backup first!
- be sure to provide “search-for” string as specific as possible, or you will get unexpected replacements (e.g. replacing mini with little will also convert all minivans into littlevans); also, do use WHERE clause when necessary to limit the number of rows modified
- the function in the example is case-sensitive, so replacing all minivans with vehicles won’t replace Minivans. However, I believe there exists a case-insensitive version of REPLACE function
Posted in how-to, Notepad | No Comments »
21st October 2009
If you get this message when opening vignettes:
Error in openPDF(vif) :
getOption(‘pdfviewer’) is ”; please use ‘options(pdfviewer=…)’
and you are tired of running this command every time:
> options(pdfviewer=”okular”)
then you should check if your system-wide Renviron file has proper PDF viewer set:
Read the rest of this entry »
Posted in *nix, how-to, Notepad, Software | No Comments »
30th September 2009
If you have happened to observe similar messages in your dmesg:
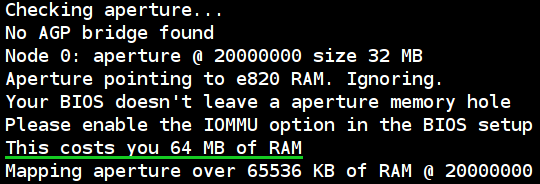
[ 0.004000] Checking aperture…
[ 0.004000] No AGP bridge found
[ 0.004000] Node 0: aperture @ 20000000 size 32 MB
[ 0.004000] Aperture pointing to e820 RAM. Ignoring.
[ 0.004000] Your BIOS doesn’t leave a aperture memory hole
[ 0.004000] Please enable the IOMMU option in the BIOS setup
[ 0.004000] This costs you 64 MB of RAM
[ 0.004000] Mapping aperture over 65536 KB of RAM @ 20000000
and you are using AMD-based system w/o AGP video, then my advice is: just leave that as is, do not bother “improving”! Any tinkering with kernel boot options won’t do you any good, as the kernel has already done the best it could.
Just a note: all those messages at the top of the post should only happen if you have 4 or more GiBs of RAM. If you have less than that, and do have those messages – my experience might be inappropriate for your case.
Another note: my BIOS does not have any IOMMU settings (or “Memory hole remapping†settings), so I didn’t try that. You should check if your BIOS has IOMMU-related options first, just as kernel message suggests.
Read on for details.
Read the rest of this entry »
Posted in *nix, how-to | No Comments »
11th June 2009
There is no way I’m aware of to do what the title says. However…
I’m sure that you are aware of the fact that floats representation in any programming language is limited by the precision of the internal binary representations. In other words, you can never have an exact float representation – there will always be some precision associated with the float you are working with. The simplest example is the difference in precision between the float and double types in C.
Suppose I have the following code fragment:
[C] if ( result.score >= input->raw_cut_off ) [/C]
Both result.score and input->raw_cut_off are of type float, and can have positive and negative values. When compared with the greater than or equal ( >= ) operator, it is not always that condition is true – for the precision reasons shortly mentioned above.
As I already said, there is no precision specification for equality operators in C. But it is quite simple to “invent” precision specification; e.g. if I wanted to test for equality only, I could write
[C] if ( fabsf( result.score – input->raw_cut_off ) < 0.000001 )[/C]
In this example, I'm effectively asking for 6-digit precision for the equality comparison of floating-point values. Note, that if you replace that 0.000001 with the actual precision limit of the floating type you are using, you will be "exactly" comparing floating-point numbers - up to that type's precision, of course  .
.
The first-most example with the >= operator can be rewritten as
[C] if ( result.score > ( input->raw_cut_off – precision) ) [/C]
where precision is exactly what it is named, e.g. precision = 0.000001.
Sources used:
Posted in how-to, Programming | No Comments »

