Overlaying gene expression data onto pathways from databases
5th November 2010
Superimposing gene expression data onto pathways from databases is a common task in the final steps of microarray data analysis – that is, biological interpretation and results discussion.
I have found many tools which claim to facilitate this procedure. Some of them are reviewed below (in no specific order).
Pathway Explorer by Bernhard Mlecnik was last updated in 2007, but is fully functional (I believe it is being maintained without changes to the last-updated date). Both online and downloadable Java applications are available. Note that for downloadable application you will need to obtain a license key – the procedure is well documented and was very fast for me.
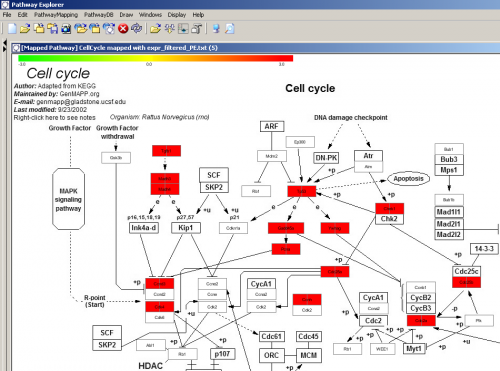
Pathway Explorer supports import from 3 sources: KEGG xml files, biocarta URLs, and GenMAPP URLs. Import from KEGG does work as described in the short manual, and seems functional (I had some problems exporting/saving the resulting picture, but didn’t investigate further). Biocarta import seems to work, but for some reason does not display expression levels of pathway components. I could not test the import of GenMAPP pathways, because they are not available online.
I found Pathway Explorer good, but then switched to PathVisio (reviewed next), because for some reason Pathway Explorer was recognising only a small fraction of genes from my expression data. It could be that identifiers mappings are outdated, but this is just a guess.
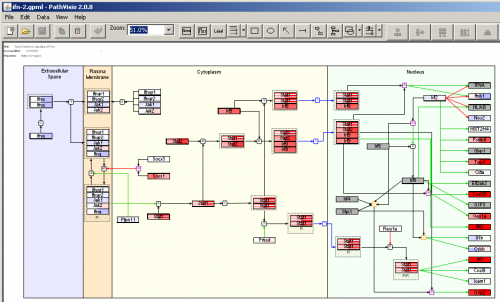
PathVisio appears to be a spin-off of GenMAPP and WikiPathways. It excells at importing/visualizing WikiPathways data, which even comes bundled with PathVisio Java application. It is easier to use than Pathway Explorer, and it seems to recognize more genes (although still not all the genes which are present in the data). There is KEGG pathways support, but it is not always usable – many edges (links between genes/proteins) are absent, so instead of a pathway you get a bunch of nodes relevant to a pathway, but cannot really see how they are connected. PathVisio supports an insanely long list of database identifiers, so it is highly unlikely that you will have to map your data to use a different identifier. This pathway mapper exports to several formats, including PNG and PDF.
I could not fully test AffyWEB, because it doesn’t list rat arrays we used. Trying their barley genome example did work, so the tool is probably functional. It overlays your expression data onto KEGG pathways.
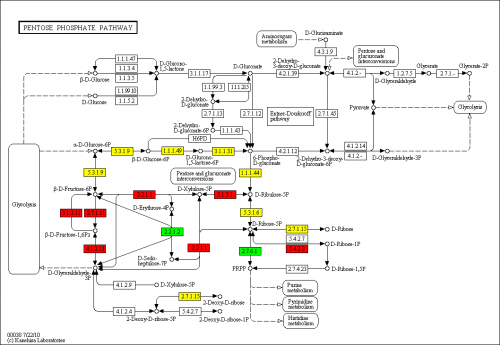
G-language Microarray System is a comparatively simple pathway visualizer. It accepts CSV files containing EntrezGene IDs column with a single column of expression values normalized to 1-100 range, fetches requested KEGG pathway, and generates a Flash (SWF) object depicting that pathway with coloured components. It does work with sample data. I was too lazy to normalize my expression data to [1;100] range, and SWF is not exactly a usable format, so I haven’t tested this tool any further (you can right-click to zoom in the flash pathway below).
If time permits (or work requires) this post may be extended with the reviews of GenMAPP, GEPAT, KEGG2heatmap script, EGAN, MapMan, Pathway Miner, ArrayXPath, VisANT, SpotXplore, and maybe others.
Please comment to share your experience using pathway expression overlaying tools or to suggest other tools.